Introns were originally thought to be ‘junk DNA’ without function but accumulating evidence has shown that they can have important functions in the regulation of gene expression. In humans and other mammals, introns can be extraordinarily large and together they account for the majority of the sequence in human protein-coding loci. However, little is known about their structural variation in human populations and the potential functional impact of this genomic variation. To address this, we have studied how copy number variants (CNVs) differentially affect exonic and intronic sequences of protein-coding genes. Using five different CNV maps, we found that CNV gains and losses are consistently underrepresented in coding regions. However, we found purely intronic losses in protein-coding genes more frequently than expected by chance, even in essential genes. Following a phylogenetic approach, we dissected how CNV losses differentially affect genes depending on their evolutionary age. Evolutionarily young genes frequently overlap with deletions that partially or entirely eliminate their coding sequence, while in evolutionary ancient genes the losses of intronic DNA are the most frequent CNV type. A detailed characterisation of these events showed that the loss of intronic sequence can be associated with significant differences in gene length and expression levels in the population. In summary, we show that genomic variation is shaping gene evolution in different ways depending on the age and function of genes. CNVs affecting introns can exert an important role in maintaining the variability of gene expression in human populations, a variability that could be related with human adaptation.
Category Archives: Papers
A new paper is out in NAR: “Automatic identification of informative regions with epigenomic changes associated to hematopoiesis”
Abstract
You can visualize the chromatin states at the UCSC browser trackhub and read the full paper here:
Automatic identification of informative regions with epigenomic changes associated to hematopoiesis, by Enrique Carrillo-de-Santa-Pau, David Juan, Vera Pancaldi, Felipe Were, Ignacio Martin-Subero, Daniel Rico and Alfonso Valencia on behalf of The BLUEPRINT Consortium
Late replicating CNVs as a source of new genes
The order in which DNA is copied during the cell division process reflects the evolutionary history of living beings: the oldest genes are copied first, and afterwards the genes that appeared later
The authors propose an original model that would explain how regions of the genome that are copied later on facilitate the birth of new genes with specific functions in tissues and organs
Press release of the work by David Juan et al (Biology Open 2013) carried out with Alfonso Valencia at CNIO.
Source: A CNIO study recreates the history of life through the genome | cnio.es
From happiness on Twitter to DNA organisation
Assortativity, a property used in network analysis, helps to identify the proteins that ensure the correct folding of DNA inside the nucleus
The new perspective allows the integration of different kinds of Big Data, unravelling the 3D configuration of the genome
Last year’s press release of the work by Vera Pancaldi et al (Genome Biology 2016) carried out with Alfonso Valencia at CNIO.
Source: From happiness on Twitter to DNA organisation | cnio.es
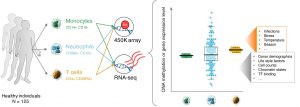
Neutrophil gene expression variability in humans
A new paper from the group is out!! A great collaboration within the BLUEPRINT Consortium, fantastic effort leaded by Simone Ecker (former PhD student at CNIO, now postdoc with Stephan Beck at UCL).
http://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1156-8