by Annie Derry
As this year comes to a close, it feels right to reflect on the accomplishments that have been made by researchers at Newcastle University (with collaboration from other institutes) this year. The academic output of the University as a whole is, of course, extremely widespread, so I will be focusing on a small fraction of the research progress made in the Medical Sciences this year.
1. Fighting against drug-resistant Tuberculosis
Tuberculosis is usually treated by the antibiotic rifampicin; however, in recent years strains of Mycobacterium tuberculosis (TB-causing bacteria) have become resistant to it, making treatment very difficult and raising concerns over potential epidemics. Tuberculosis can be fatal without treatment and drug-resistant strains may become more prominent, leading to increased deaths worldwide. Newcastle University and Demuris Ltd researchers have made a promising discovery in a naturally occurring antibiotic called kanglemycin A. This antibiotic works in a similar way to rifampicin but has been identified to be active in working against Mycobacterium tuberculosis that are resistant to rifampicin. This is due to kanglemycin A’s ability to bind with stronger affinity to the RNA polymerase molecules of the resistant strain. This discovery can be used in drug development in the future in order to prevent any further fatalities from rifampicin-resistant tuberculosis.
Read the full article by Mosaei, Molodtsov, and colleagues
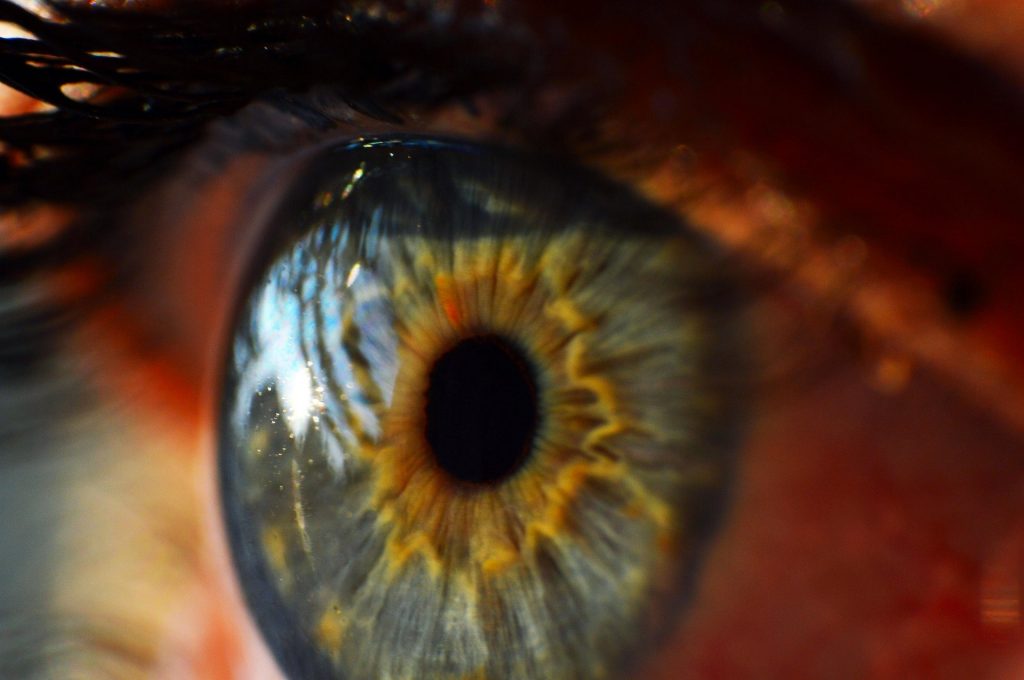
2. The first 3D-printed human cornea
The cornea is a fundamental part of the eye which acts as the outer-most lens, while preventing microbes, dirt and dust from entering. The cornea can become damaged due to injury, infection or conditions that cause corneal swelling, such as Fuchs’ endothelial dystrophy. There is often a shortage of corneas available for transplantation in people who need them, meaning that patients suffer visual impairment and potential blindness. These scientists at Newcastle University have devised a technique that uses stem cells from the corneas of healthy donors to create a “bio-ink”. This ink is then used in a 3D bio-printer to form the shape of a human cornea in under 6 minutes! The stem cells then use this as a template to grow into a cornea that can be used for transplantation. Although this method is a while away from being available to the general public it is an extremely exciting and influential development!
Read the full article by Isaacson, Swioklo, & Connon
3. A new link between Tuberculosis and Parkinson’s disease
This collaborative study led by Newcastle University, the Francis Crick Institute and GSK suggests that drugs we design for Parkinson’s disease might actually be able to treat tuberculosis as well. At the moment the focus for Parkinson’s treatment is to make drugs that block LRRK2, a protein which becomes overactive in the disease due to mutations in the LRRK2 gene. The results from this study indicate that this LRRK2 protein prevents the clearance of Mycobacterium tuberculosis by stopping phagosomes fusing with lysosomes in the immune response. Therefore, a drug that blocks LRRK2 protein could potentially be useful in helping the immune system to fight a tuberculosis infection.
Read the full article by Härtlova, Herbst, and colleagues
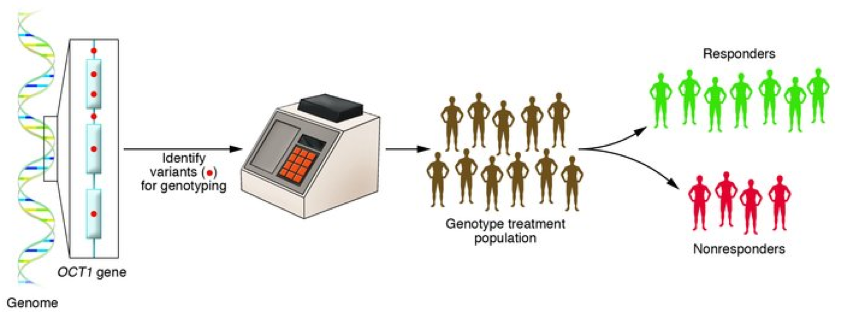
4. Further insights into reversing Type 2 diabetes
This study, funded by Diabetes UK, set out to decipher why some patients with Type 2 diabetes can recover to regular health through weight loss, and why other patients cannot. The study used data from a subset of individuals already taking part in the DiRECT (Diabetes Remission Clinical Trial) study which used non-surgical weight management to see whether it would lead remission of the disease. The researchers at Newcastle saw that there was one main difference between those who responded to the weight loss (and entered remission) and those who did not. The ‘responders’ had insulin-producing beta cells that appeared to start secreting the correct amount of insulin again after losing weight, while ‘non-responders’ had no change in insulin production despite weight loss. Responders were generally found to have lived with Type 2 diabetes for slightly less time on average than non-responders, which suggests that if beta cells are not under stress from increased fatty deposits for too long, they may still be able to recover when fat loss occurs.
Read the full article by Taylor, Al-Mrabeh, and colleagues.
5. Improvements in treatment for bone marrow cancer
Myeloma is a type of blood cancer that develops from plasma cells within the bone marrow. It has a 5-year survival rate of 50% and there is no complete cure, but there are ways to treat and manage the cancer in order to increase the amount of time patients survive for. In a study published by The Lancet Oncology, researchers at Newcastle and Leeds University monitored over 4000 patients in the UK for 7 years, in order to assess the efficacy of the drug lenalidomide (not yet available on the NHS). The patients had already completed their initial treatment and some were then randomly chosen to use lenalidomide during drug therapy. The results showed that lenalidomide significantly extended the lives of the patients and it reduced the risk of cancer progression or death by more than 50%. This study therefore shows a very promising new treatment method for people living with myeloma, which hopefully will be seen within the NHS before too long.
Read the full article by Jackson, Davies and colleagues
These publications are representative of just a tiny amount of the work at Newcastle University that has had global success this year, and some additional links are listed below. Look out for more North East research developments in 2019 – happy New Year!
——–
If you would like to learn more about life science research news from Newcastle University please follow the links: https://www.ncl.ac.uk/medicalsciences/news/ & https://www.ncl.ac.uk/press/archive/
Breakthrough in childhood brain cancer
Tobias Goschzik PhD, Edward C Schwalbe PhD, Debbie Hicks PhD, Amanda Smith MSc, Anja zur Muehlen, Prof Dominique Figarella-Branger MD et al.
Potential to use gene-editing to halt kidney disease
Simon A. Ramsbottom, Elisa Molinari, Shalabh Srivastava, Flora Silberman, Charline Henry, Sumaya Alkanderi, Laura A. Devlin, Kathryn White, David H. Steel, Sophie Saunier, Colin G. Miles, and John A. Sayer
A breast cancer drug that could be used to treat leukaemia
Natalia Martinez-Soria, Lynsey McKenzie, Julia Draper, Georges Lacaud, Constanze Bonifer, Olaf Heidenreich et al.
Stalling the cell cycle to put cancer cells to sleep
Teemu P. Miettinen, Julien Peltier, Anetta Härtlova, Marek Gierliński, Valerie M. Jansen, Matthias Trost and Mikael Björklund.