By Rosie Gathercole
Working with poo turned out to be exactly the summer experience I wanted!
I worked at the national Cryptosporidium Reference Unit (CRU) at Public Health Wales in Swansea with Professor Rachel Chalmers and her team. I received a Scholarship from the Society for Applied Microbiology (SfAM) for this placement, writing the application together with Rachel.

Cryptosporidium is a parasite that causes diarrhoea, is found globally and is typically passed from animals, other people, food and fresh water sources. It is currently a human health issue due to the significant effect it has in developing countries and the lack of specific treatments to fight the parasite. Quite often how well you recover from the illness depends on how healthy you were to begin with!
What did I actually do in Swansea?
After reviewing the literature, we quickly realised that it would be very useful if we could analyse the protein content of Cryptosporidium because a clear understanding of proteomics can help the current genomics work being done in the area. To do this the parasites would first have to be isolated from human faecal samples at the CRU.
“The CRU gets sent lots of samples, yes that means lots of parasite filled poo!”
My project was to optimise a way to isolate parasite “eggs” from these poo samples ready for protein analysis. What I ended up doing was performing ‘salt flotations’ initially to separate the parasites from the faeces. This involved placing the samples on salt-water and spinning in a centrifuge. Next, I used potassium bromide solutions to separate contents specifically based on their density. However, the more you try and ‘clean-up’ a sample, the more parasites you lose from it so it became a bit of a balancing act which involved fiddling with centrifuge speeds, volumes of samples, and reagents for a week or two.
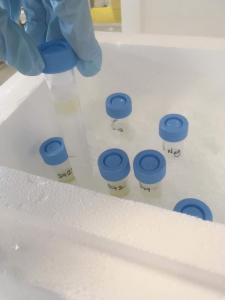
This photo shows the samples (still a lovely brown colour) on top of the potassium bromide solutions. After spinning you can now see a white band containing the parasite eggs. This band was then extracted and washed.

It worked!
In order to count the number of eggs that I could purify using this method, I pipetted the extracted samples onto a tiny grid on a microscope slide. I was hoping to be able to purify at least 1 million eggs in order to have sufficient material for the next step so I was ecstatic to come across a sample with over 10 million!

Each “egg” that can be seen contains four Cryptosporidium parasites, so my sample, actually contained more than 40 million parasites. All purified in around 1ml of clear liquid.
After collecting as many good samples as I could, I presented my protocol and results to the CRU team to show them that I had been successful since letting me loose in their lab.
Analysing my purified parasites
Next, I suspended the parasites in ethanol to kill any contaminants, packaged them in dry ice, and drove to Liverpool University Veterinary School where I was lucky enough to work with Erika Transfield, a MALDI application specialist for the Bruker Corporation.

My aim at Liverpool was to analyse my purified Cryptosporidium samples using their latest MALDI-TOF mass spectrometer. Mass spectrometry machines are able to tell you what the mass for every bit of protein in your sample and from this you can determine which proteins are present. This had never been performed successfully with Cryptosporidium before, but we were very hopeful that the developed protocol would be successful.
First, I added a bunch of chemicals to my samples to “fix” the proteins and then I pipetted them onto a metal plate that would go into the machine.

Success!
What you can see on the computer screen behind me was the first successful protein peaks (spectrum) that the computer spat out. After a few hours we ended up with two successful spectra from the six samples that I had brought with me from Swansea. Despite being back at university for my final year, I am still in contact with the team as we analyse the data to identify the specific protein fragments the mass spectrometer identified.

On to Aberystwyth University
Straight after generating this exciting data, I then was lucky enough to be whisked away to Aberystwyth University where I spent two more weeks in the bioinformatics office with a PhD student who showed me how to work with massive data sets. We used operating systems such as Linux to analyse the complex data generated from these parasites. We used scripts to see if any of my samples were unique e.g. If one sample contains proteins that none of the others did, that could be a way to identify that subtype of Cryptosporidium?
….and finally
” I would definitely recommend doing a placement over the summer”
This experience as a whole was able to provide me with a seemingly endless list of transferable skills which I am already finding to have transformed the way I approach my 3rd year studies. Also being able to work so freely within a research-based work environment has fuelled my interest in lab-based science and is certainly influencing my post-graduation plans. Being able to work side-by-side with a talented bioinformatician in particular has kick-started my passion for the field and I am still continuing to play with the software I was introduced to just a few months ago.
I would definitely recommend doing a placement over the summer and I am incredibly grateful to Professor Rachel Chalmers for taking the time to organise and oversee my project, her team for helping me in the lab and SfAM for the funding.
Rosie did this fantastic work in the summer between her second and final year of BSc Biomedical Science in the School of Biomedical, Nutritional and Sport Sciences.
Read about summer placements our other students have done here.

