“Most of us know that we should wash our hands after being around animals but do most of us know the reasons why? As a researcher who spends most of his time in the lab killing E. coli, using E. coli specific antibiotics I should be well aware of the dangers of this often underestimated Gram-negative bacteria. However when my youngest daughter contracted E. coli O157 after visiting an agricultural show in Scotland in 2011 (even though we had followed the hand washing procedure), I realised that I did not appreciate how nasty this bacteria can be. Thankfully after 6 weeks in hospital including a lengthy stretch on dialysis my daughter made a full recovery but not everyone is so lucky.”
The urgent need to develop new drugs to target pathogenic bacteria has been a theme of the ICaMB blog since its inception in early 2013. However, these words from Dr Chris Johnson, a postdoc in Jeremy Lakey’s laboratory, bring home the seriousness of the problem. Fortunately, Chris is in a position to do something about this.
Many of you will have realised that last week saw another potential breakthrough in ICaMB’s ongoing ‘War on bacteria’ (and here, here and here). This time the PI making the news, with data demonstrating the possibility of a whole new class of antibiotics, was Professor Jeremy Lakey. And when we say making the news, we mean that literally. The ITV interview of Jeremy and his team can be viewed here
This story has made news around the world and has been featured in newspapers in Australia and India (and here), as well as closer to home. Here is the official university press release.
So what’s going on? If you want to read the primary paper it is here
The unstructured domain of colicin N kills Escherichia coli. Mol Micro 89:84-95
Chris Johnson, who was the lead author on this manuscript explains:
“E. coli produces protein antibiotics called colicins which are used to kill E. coli and closely related bacteria in the eternal bacterial arms race. In order to further understand how one of these, colicin N (ColN), works, we dissected the protein into its individual domains to see how each part behaved in isolation. Quite by chance we found that part of the protein, THE TAIL, was actually toxic to them. Although far less efficient than the entire ColN molecule, it remains specific for E. coli and furthermore, the specificity is housed within an intrinsically unfolded domain (a domain which has no defined 3D structure). Although this is a very basic discovery in its early stages, it allows us to appreciate novel mechanisms to kill E. coli.” (see the full version of this at the bottom of the page)
Some more about Jeremy
 Any of you who were bioscience undergraduates in Newcastle will know Jeremy Lakey from his famous recreations of protein structure using party balloons. Others may know him for co-founding Orla Protein technologies. Some of us know him as a man who will always buy his round in the pub. All of us know him
Any of you who were bioscience undergraduates in Newcastle will know Jeremy Lakey from his famous recreations of protein structure using party balloons. Others may know him for co-founding Orla Protein technologies. Some of us know him as a man who will always buy his round in the pub. All of us know him  as a great scientist and colleague. Furthermore, Jeremy is a leading supporter of Leading Edge and recently a group of 6 Year 9 school pupils from St Cuthberts RC School looked at how ColN acts against E.coli when you start changing the amount of salt they are grown in. As many ICaMB scientists may know Jeremy also runs a workshop with Ponteland Community High School to explore bacterial shape and their surfaces using LEGO.
as a great scientist and colleague. Furthermore, Jeremy is a leading supporter of Leading Edge and recently a group of 6 Year 9 school pupils from St Cuthberts RC School looked at how ColN acts against E.coli when you start changing the amount of salt they are grown in. As many ICaMB scientists may know Jeremy also runs a workshop with Ponteland Community High School to explore bacterial shape and their surfaces using LEGO.
But did any of us think that one day Jeremy Lakey may SAVE THE WORLD from antibiotic resistant bacteria? Possibly.
The detailed science
Most Gram- negative bacteria produce protein antibiotics which are used as weapons in the battle between competing populations of bacteria. E. coli produces protein antibiotics called colicins which are use to kill E. coli and closely related bacteria. Once colicins are released into the extracellular milleu they dock onto their targets via specific outer-membrane receptors and then seek out an internal, periplasmic, binding partner (the Tol or Ton proteins) which helps them translocate into the cell. We study colicin N (ColN) which comprises of an intrinsically unfolded N-terminal translocation (T) domain, involved in TolA and OmpF binding. Its central receptor binding (R) domain binds lipopolysaccharide whilst its C-terminal 200 amino acids define the cytotoxic pore-forming (P) domain. This latter feature is common to all pore-forming colicins and forms a channel in the inner-membrane causing K+ release and cell death. Other colicins have C-terminal domains which display cytotoxic activities that include DNAase or RNase activity. Irrespective of the particular cytotoxic activity, all colicins are comprised of three domains (T-R-P) and it was assumed that the sole role of the T and R domains was to deliver the cytotoxic C-terminal domain across the outer-membrane.
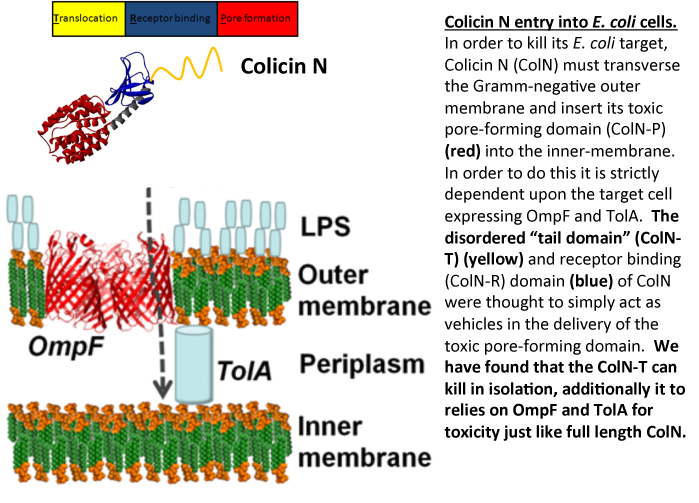 In order to investigate the mechanism of ColN activity we dissected the protein into its individual domains. We were attempting to block the toxic activity of full length ColN by pre- incubating E. coli cells with the intrinsically unfolded T-domain. The rationale behind the experiment was that we could block all the available receptor sites on the E. coli target cells by saturating with T-domain, such that when the full length ColN was added to cells it would be non-toxic, as all the essential receptor binding sites would be already sequestered. However rather than protecting the cells, T-domain was found to be toxic and like full length ColN provoked K+ efflux. Although less efficient than full length ColN, T-domain is strictly dependent upon the same receptor proteins, OmpF and TolA for killing. Since these receptors are only found in E. coli-like bacteria, T-domain displays the unusual combination of a generic killing mechanism coupled with extreme specificity housed within an intrinsically unfolded domain.
In order to investigate the mechanism of ColN activity we dissected the protein into its individual domains. We were attempting to block the toxic activity of full length ColN by pre- incubating E. coli cells with the intrinsically unfolded T-domain. The rationale behind the experiment was that we could block all the available receptor sites on the E. coli target cells by saturating with T-domain, such that when the full length ColN was added to cells it would be non-toxic, as all the essential receptor binding sites would be already sequestered. However rather than protecting the cells, T-domain was found to be toxic and like full length ColN provoked K+ efflux. Although less efficient than full length ColN, T-domain is strictly dependent upon the same receptor proteins, OmpF and TolA for killing. Since these receptors are only found in E. coli-like bacteria, T-domain displays the unusual combination of a generic killing mechanism coupled with extreme specificity housed within an intrinsically unfolded domain.
Links
Jeremy Lakey’s University home page: http://www.ncl.ac.uk/camb/staff/profile/jeremy.lakey
Follow Jeremy Lakey on Twitter: https://twitter.com/JeremyLakey
ICaMB: http://www.ncl.ac.uk/camb/
The Centre for Bacterial cell Biology: http://www.ncl.ac.uk/cbcb/
The official Newcastle University press release: http://www.ncl.ac.uk/press.office/press.release/item/chance-finding-could-lead-to-new-antibiotics
Link to ITV story:
http://www.itv.com/news/tyne-tees/story/2013-07-05/breakthrough-in-combatting-bacterial-infection/
The ‘Australian’ story: http://www.theaustralian.com.au/news/breaking-news/tail-could-be-used-for-new-drugs/story-fn3dxix6-1226674131684
The ‘Times of India’ Story: http://articles.timesofindia.indiatimes.com/2013-07-06/science/40406695_1_escherichia-coli-e-coli-protein
The Northern Echo story: http://www.thenorthernecho.co.uk/news/10524003.Scientists__chance_find_may_develop_new_generation_of_antibiotics/
Jeremy’s company, Orla Proteins: http://www.orlaproteins.com/about-orla/the-board.aspx