In this week’s blog, Professor Martin Embley reflects on the the journey that led to him, his collaborators and his laboratory to fundamentally change our views on evolution and the Tree of Life.
In this week’s blog, Professor Martin Embley reflects on the the journey that led to him, his collaborators and his laboratory to fundamentally change our views on evolution and the Tree of Life.
The Early Years
My early career was a bit of a random walk while I tried to figure out what I really wanted to do. After my PhD in Newcastle on bacterial diseases of trout and salmon, I got a job teaching industrial microbiology at North East London Polytechnic in 1984. It was an odd but interesting place, a number of staff appeared to have strong religious beliefs of various sorts and wanted to talk about them, and one colleague thought he could change traffic lights from red to green so he never had to slow down. I was keen to keep doing some research and I was interested in evolution, but like a lot of newly independent researchers I struggled to get any funding. My big break came when I got a “cultural exchange” grant from Newham Council to go to Poland to learn some molecular biology and I met Professor Erko Stackebrandt who was passing through. Erko had worked with Carl Woese in pioneering the use of ribosomal RNA sequences to investigate evolutionary relationships among prokaryotes. I persuaded him to let me visit his lab in Germany to learn the new techniques and in 1991 these skills got me a job at the Natural History Museum in London.
The Museum wanted to set up a lab using molecular sequences to investigate evolutionary relationships. The film Jurassic park was just about to appear and there was tremendous excitement about the potential of ancient DNA. The Museum gave me free rein regarding my own research as long as it had evolution at its core. So I decided to work on the early evolution of eukaryotic cells. At the time two ideas were central to views of early eukaryotic evolution. One was that the “three domains tree of life” was an accurate description of the relationships between eukaryotes and prokaryotes (you can see the tree here). The other was that some eukaryotes, including obligate intracellular microsporidian pathogens, had never had mitochondria because they split from other eukaryotes before the mitochondrial endosymbiosis. I’ve been trying to test these two ideas for the past 25 years and while it’s often been difficult and frustrating, it has also been a lot of fun.
to work on the early evolution of eukaryotic cells. At the time two ideas were central to views of early eukaryotic evolution. One was that the “three domains tree of life” was an accurate description of the relationships between eukaryotes and prokaryotes (you can see the tree here). The other was that some eukaryotes, including obligate intracellular microsporidian pathogens, had never had mitochondria because they split from other eukaryotes before the mitochondrial endosymbiosis. I’ve been trying to test these two ideas for the past 25 years and while it’s often been difficult and frustrating, it has also been a lot of fun.
A Team Effort
Like most PI’s I’ve relied on attracting talented young scientists to do the work that we have published. Robert Hirt walked into my lab off the street and asked me if he could do a project involving eukaryotic evolution and ecology. He already had a first author paper in Cell and now he wanted to do something different. We didn’t do much ecolog y together but Robert and I co-supervised PhD student Bryony Williams who showed that microsporidians actually contained a tiny, hitherto overlooked mitochondrion, now often called a mitosome.
y together but Robert and I co-supervised PhD student Bryony Williams who showed that microsporidians actually contained a tiny, hitherto overlooked mitochondrion, now often called a mitosome.
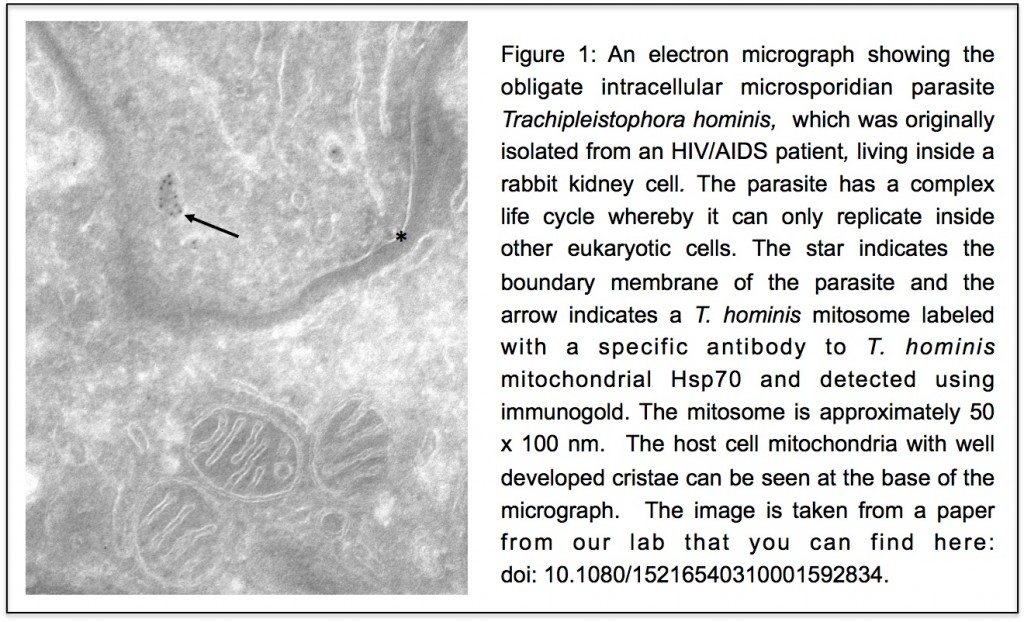
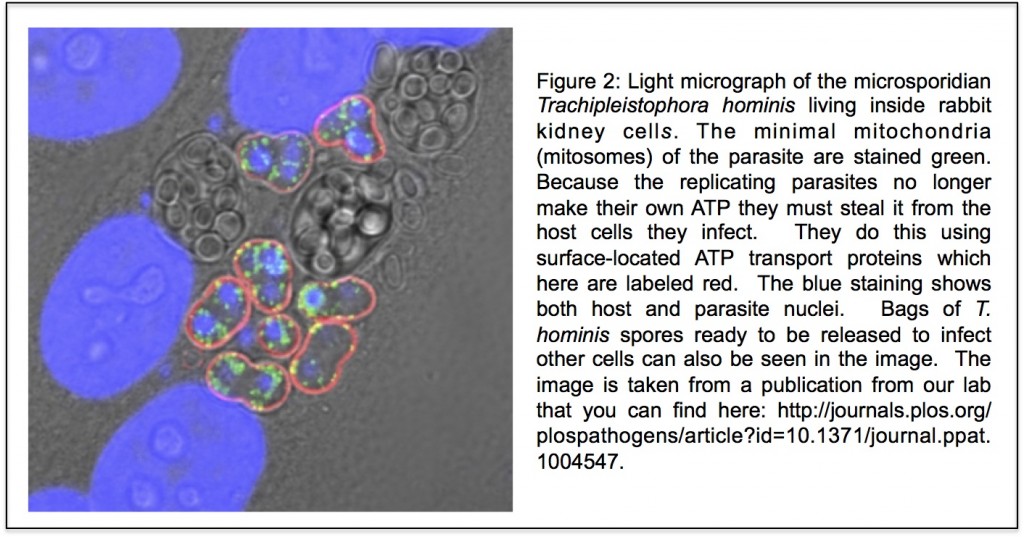
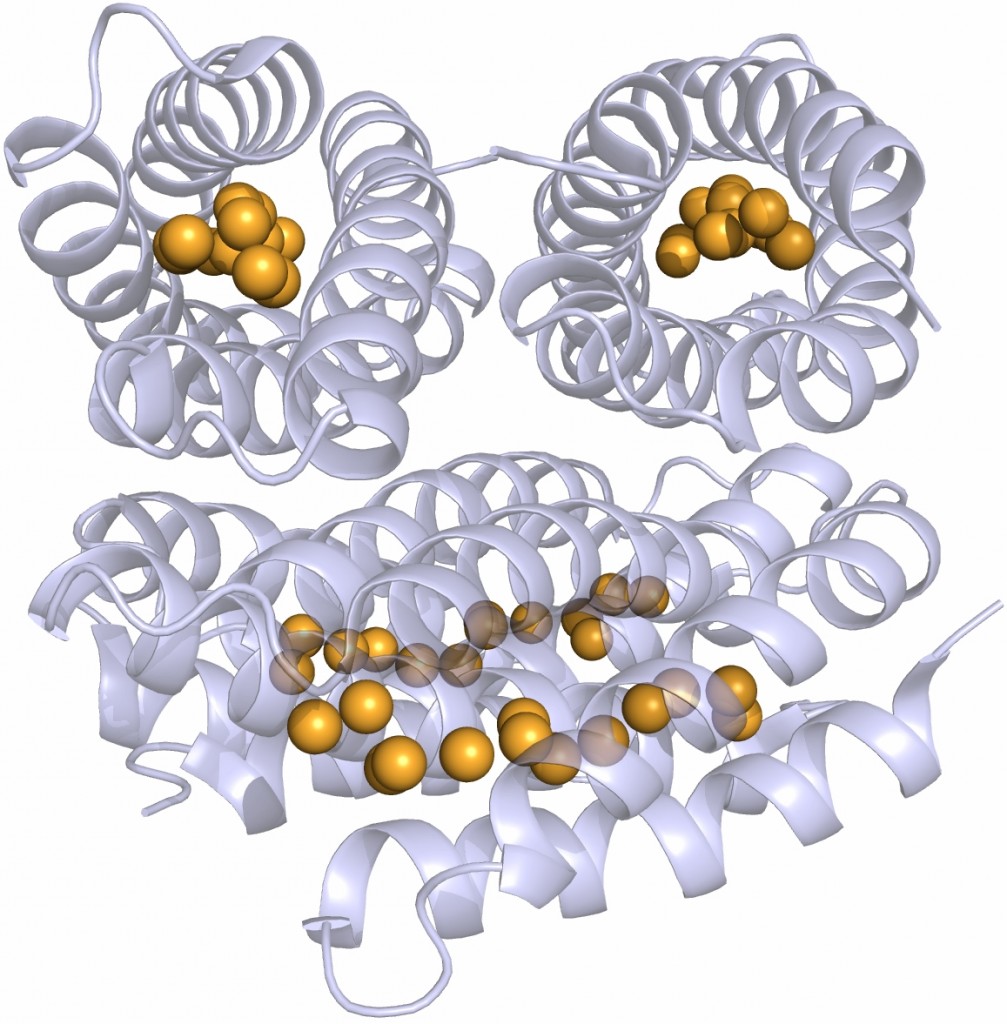
Unlike our own mitochondria, the microsporidian mitosome doesn’t make ATP, because it has lost all of the pathways used by classical mitochondria to make energy. Alina Goldberg in our lab – now at Newcastle – and Sabine Molik in the lab of Roland Lill in Germany spent  the next seven years showing that the mitosome functions in the biosynthesis of essential cytosolic and nuclear Iron/Sulphur (Fe/S) proteins. The discovery of a tiny mitochondrion in microsporidia (Figures 1 and 2) was an important piece of evidence that led to current ideas that the mitochondrial endosymbiosis occurred at the origin of eukaryotes. Thus, it is now thought that all eukaryotes contain an organelle related to mitochondria, and its most conserved function is in Fe/S protein biogenesis, not ATP production.
the next seven years showing that the mitosome functions in the biosynthesis of essential cytosolic and nuclear Iron/Sulphur (Fe/S) proteins. The discovery of a tiny mitochondrion in microsporidia (Figures 1 and 2) was an important piece of evidence that led to current ideas that the mitochondrial endosymbiosis occurred at the origin of eukaryotes. Thus, it is now thought that all eukaryotes contain an organelle related to mitochondria, and its most conserved function is in Fe/S protein biogenesis, not ATP production.
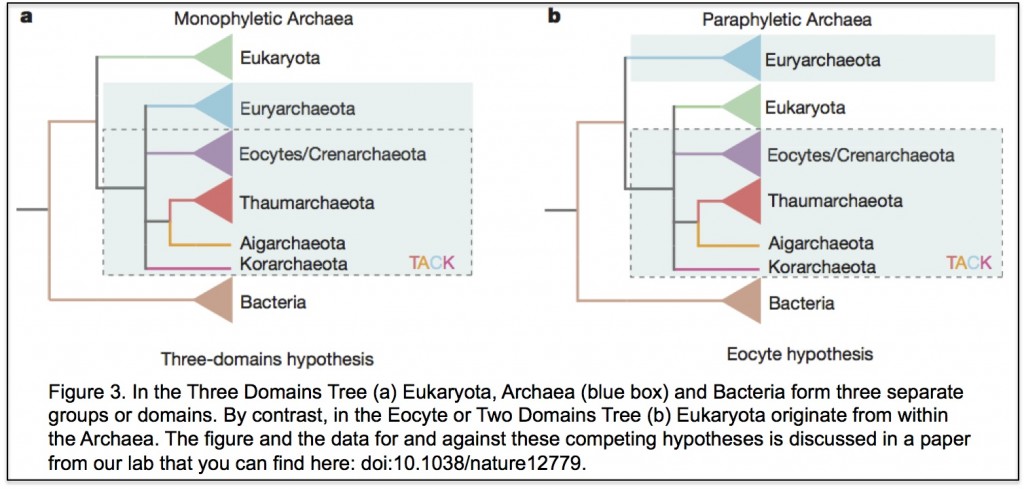
In the three domains tree of life, eukaryotes are a separate domain that is most closely related to the domain Archaea and the host for the mitochondrial endosymbiont is already a eukaryote. Although this hypothesis appears in most textbooks, there have actually been a number of alternative hypotheses published over the years (Figure 3 shows one of them), but these have largely been ignored. Cymon Cox spent three years analysing molecular sequence data to identify which of the competing published hypotheses was best supported and reached the surprising conclusion that it was not the three domains tree but an alternative hypothesis called the eocyte tree (you can read a discussion about the differences between the two trees including a picture of the eocyte tree here). In the eocyte tree, eukaryotes originate from within the Archaea, suggesting that eukaryotes are not a primary domain of life like Archaea and Bacteria, but are instead a product of genetic and cellular contributions from both prokaryote domains. Very excited by these results, we sent Cymon’s paper to Nature where it was reviewed and quickly rejected. We appealed, it was revised, it was reviewed again, and it was rejected again, all in all pretty dispiriting, but a common experience for most scientists. However, after hearing me talk about Cymon’s work at a meeting, we were invited to submit Cymon’s paper to PNAS where it was finally published in 2008.
Although Cymon’s paper has been highly cited it is true to say that the initial response from the community was very mixed. We received emails suggesting that we had manipulated our results to get the answer we wanted and one of the reviewers told us that it was impossible to infer such ancient events using molecular sequences. In responding we agreed that it was difficult to be confident about anything that happened billions of years ago based upon small amounts of data and even the best methods of analysis, but that people in the field seemed happy to use the same data and worse methods to support the three domains tree. Cymon eventually moved on, scarred but not defeated, and Tom Williams took over the project when he came to Newcastle on a Marie Curie Fellowship in late 2010. Over the next few years, more data was published as new Archaea were discovered and new methods of analysis were developed, and every analysis that Tom, or others, did on these data produced a version of the eocyte tree, so that it is now the best supported hypothesis – at least in our opinion.
More evidence emerges
Hypotheses are only useful when they make predictions that can be tested by further research, and evolutionary hypotheses are no different. The eocyte tree predicts that new species that share more features in common with eukaryotes will be discovered among the Archaea, and this prediction now appears to have been spectacularly fulfilled by recent discoveries from Thijs Ettema’s lab in Sweden. The new paper describes the discovery, so far only from metagenome data, of an archaeal lineage called Lokiarchaeota that contains many genes for proteins that were previously thought to be eukaryotic specific, including homologues of proteins used in the eukaryotic cytoskeleton, in membrane remodeling and in phagocytosis. This is incredibly exciting and the challenge is now to isolate Lokiarchaeota and other new lineages into culture so that their biology and physiology can be studied in the laboratory.
An Interesting Journey
Scientific work is often written up as if it were a linear progression towards improved understanding, a type of “Whig history” which does not accurately reflect how science is really done. In reality, science is a collaborative endeavour with lots of dead ends, confusion and false trails, and we could easily be walking down some of those still. Nevertheless, the currently prevailing paradigm for eukaryotic evolution is now very different to the popular views held in the 1990s when I started my research career. All eukaryotes are now thought to contain a mitochondrial homologue that generally functions in Fe/S protein biogenesis, and the host for the mitochondrial endosymbiont is thought to have originated from within the Archaea. Eukaryotes are thus viewed as the product of an interaction between (at least) those two prokaryotic partners and are not a primary domain of life but one derived from prokaryotic antecedents. The complex features that we take to define eukaryotic cells including our own, such as the nucleus, large genomes and diversity of RNAs, are thus secondary features that have evolved since those primordial interactions. I’m not sure what my religious former colleagues would have made of the work I’ve done since leaving NELP, but it’s been an enjoyable and interesting journey for me.
Links
The Tree of Life: http://www.pnas.org/content/87/12/4576.full.pdf
Bryony Williams’ paper: http://www.nature.com/nature/journal/v418/n6900/full/nature00949.html
Alina Goldberg and Saline Molik paper: http://www.nature.com/nature/journal/v452/n7187/full/nature06606.html
Mitochondria and Fe/S proteins: http://www.nature.com/nature/journal/v440/n7084/abs/nature04546.html
The eocyte tree: http://phenomena.nationalgeographic.com/2012/12/20/redrawing-the-tree-of-life/
Cymon Cox paper: http://www.pnas.org/content/105/51/20356.full
Tom WIlliams paper: http://www.nature.com/nature/journal/v504/n7479/full/nature12779.html
Thijs Ettema lab paper: http://www.nature.com/nature/journal/v521/n7551/abs/nature14447.html

 Last year we brought you details of the inaugural
Last year we brought you details of the inaugural