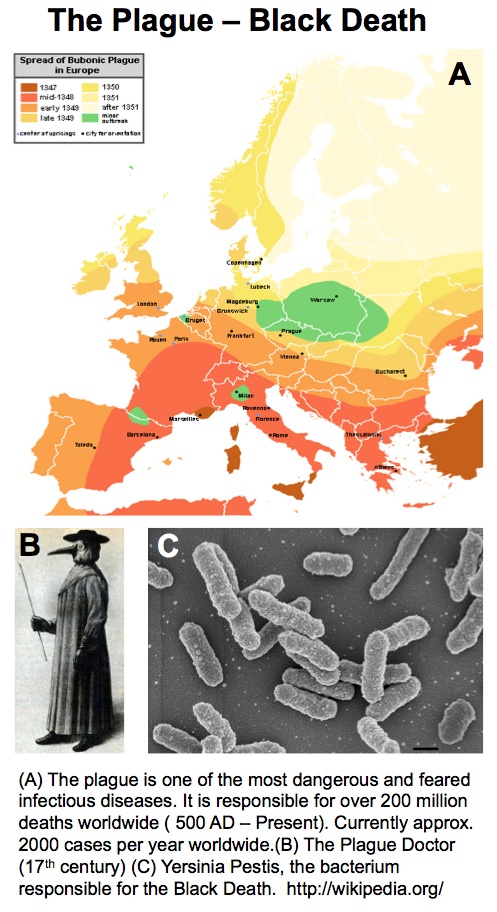
UK Government Minister Vince Cable recently announced the results of the first round in the BBSRC-supported Industrial Biotechnology Catalyst scheme where £20 M was distributed across 23 projects. Here, ICaMB’s Jeremy Lakey describes the curious scientific route that led from researching Yersinia pestis, the bacteria responsible for the bubonic plague, to a potential biotechnology breakthrough.
UK Government Minister Vince Cable recently announced the results of the first round in the BBSRC-supported Industrial Biotechnology Catalyst scheme where £20 M was distributed across 23 projects. Here, ICaMB’s Jeremy Lakey describes the curious scientific route that led from researching Yersinia pestis, the bacteria responsible for the bubonic plague, to a potential biotechnology breakthrough.
By Professor Jeremy Lakey
The project that I put together with Neil Perkins (ICaMB), Dave Fulton (Chemistry), Matt German (Dentistry) and Nick Reynolds (Dermatology), called (rather snappily I think) Manufacture of complex protein polymers for industry and medicine, is one of the recently announced Industrial Biotechnology Catalyst awards. It’s a £2.4 million research programme with a BBSRC 80% contribution of £1.8 million and quite honestly a year ago I’d never imagined having this amazing five years funding to realise this project that has nagged at me for at least the last five years.
The early years
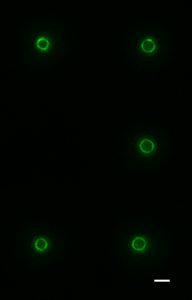
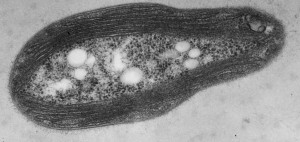
 My interest in the Caf1 protein first arose from a project with the Defence Science and Technology Laboratory (DSTL) on vaccines against possible bioweapons such as anthrax and plague. Other labs had shown Caf1 to be a chaperone usher (CU) family protein, secreted through the outer membrane of the plague bacterium Y. pestis as non-covalently linked polymers. However, most members of this family were visible under the electron microscope (EM) and had a defined structure. We tried in vain to see it under the EM for a couple of years and had given up but, as luck would have it, the world authority on EM observation of proteins using negative stain, Robin Harris, had retired from his job in Germany and moved to Hexham a town in the Tyne Valley near Newcastle. He agreed to have a look with my student at the time, Andrei Soliakov. By adding very low concentrations of protein and using his magic staining recipe, they got amazing images on the first day (see figure). I was on the other end of a country modem at my brother’s farm and so for me the images unfolded slowly down the screen. Robin, Andrei and I (much later in the day), were thus the first folk ever to see one of the key proteins of the Black Death which, between 1346 and 1353, killed up to half of the population of Europe. It was one of those sobering moments in life.
My interest in the Caf1 protein first arose from a project with the Defence Science and Technology Laboratory (DSTL) on vaccines against possible bioweapons such as anthrax and plague. Other labs had shown Caf1 to be a chaperone usher (CU) family protein, secreted through the outer membrane of the plague bacterium Y. pestis as non-covalently linked polymers. However, most members of this family were visible under the electron microscope (EM) and had a defined structure. We tried in vain to see it under the EM for a couple of years and had given up but, as luck would have it, the world authority on EM observation of proteins using negative stain, Robin Harris, had retired from his job in Germany and moved to Hexham a town in the Tyne Valley near Newcastle. He agreed to have a look with my student at the time, Andrei Soliakov. By adding very low concentrations of protein and using his magic staining recipe, they got amazing images on the first day (see figure). I was on the other end of a country modem at my brother’s farm and so for me the images unfolded slowly down the screen. Robin, Andrei and I (much later in the day), were thus the first folk ever to see one of the key proteins of the Black Death which, between 1346 and 1353, killed up to half of the population of Europe. It was one of those sobering moments in life.
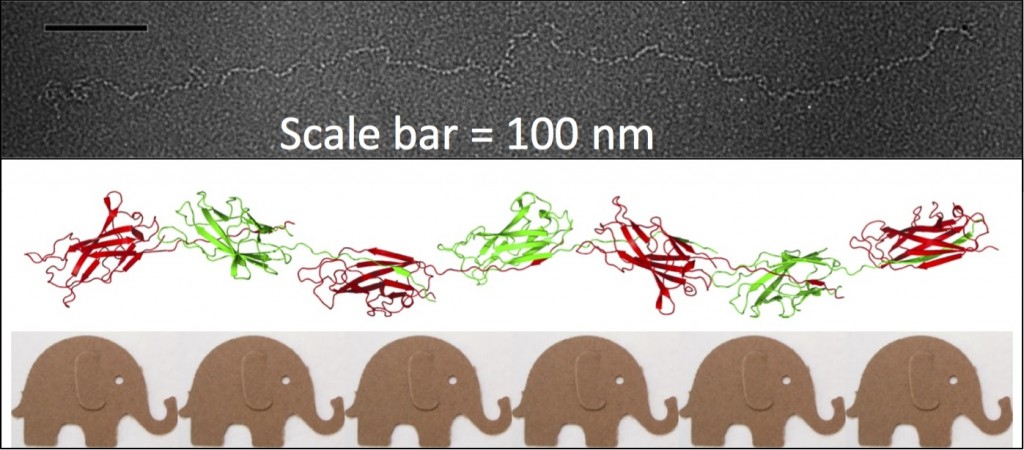
The Caf1 polymer as seen using an electron microscope (top). The structure (middle) resembles a line of circus elephants (bottom).
We also imaged these amazing molecules on the vaccine adjuvant, they looked like Christmas paper chains hanging off the sides of the adjuvant particles. No one had seen Caf1 or proteins on adjuvants before, even though most of us have been vaccinated using this material. I was swept away by the sheer coolness of the data we had and submitted a paper to Nature, then Science, then somewhere else and somewhere else after that but it soon appeared no one was quite as amazed as I was. This remains true to this day with the eventual paper in Vaccine (Ref 1 below) still only having 2 non self- citations; something I still fail to understand.
It’s love actually
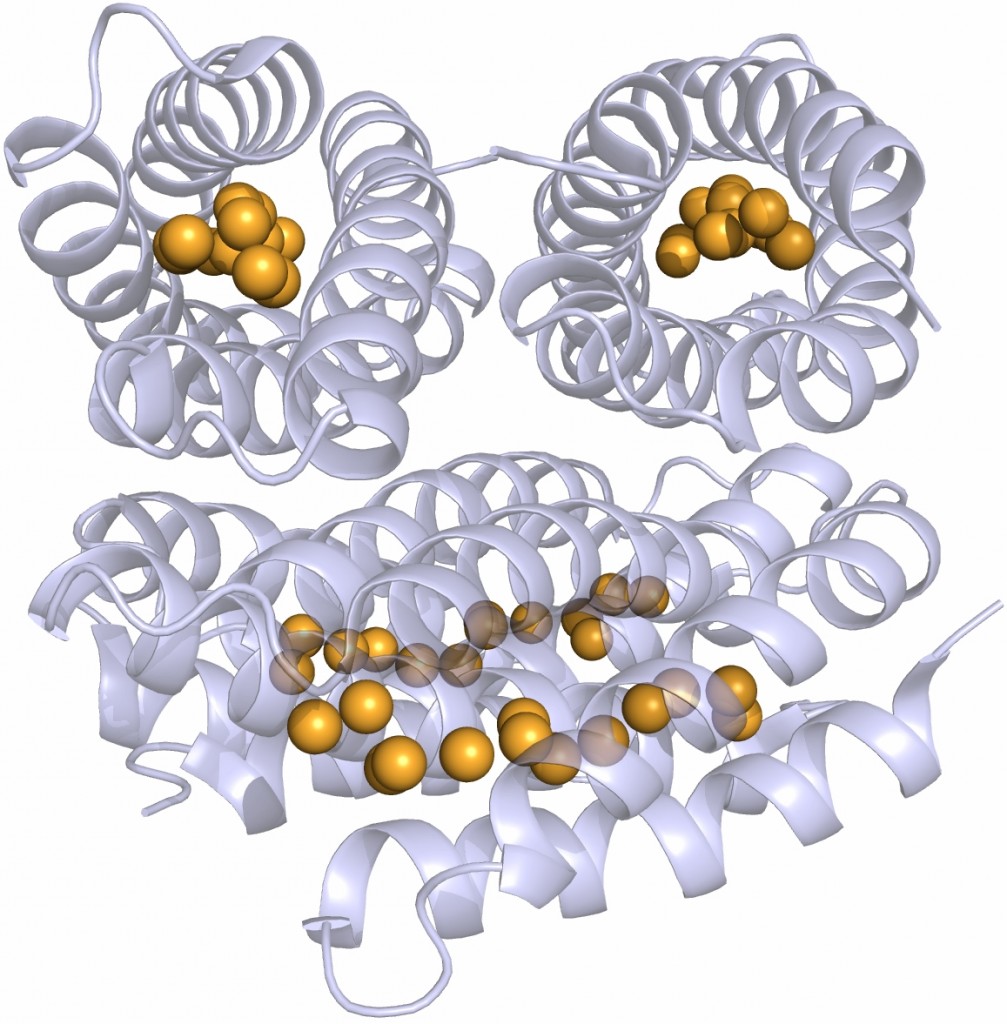
However I was seriously smitten by this molecule and hardly cared. This infatuation was made worse one day when, in a seminar, somebody showed the structure of the extracellular matrix protein fibronectin or at least the domains around the integrin binding sites where the well-known RGDS sequence motif is situated. These protein domains looked just like Caf1 because they were both immunoglobulin-like domains.
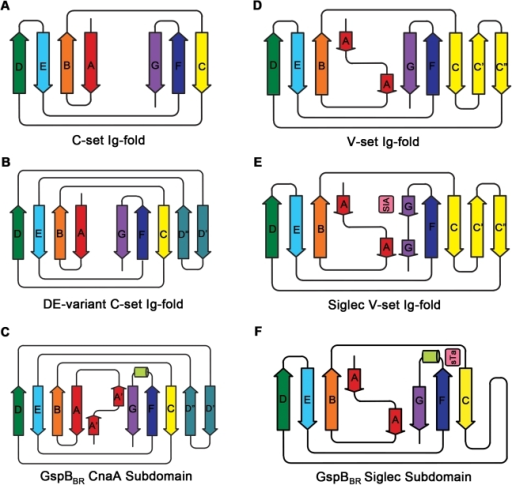
Different immunoglobulin-like protein topologies (from Pyburn et al, PLoS Pathog. 2011)
Of course, I should have seen this before as I go on and on and on about protein families in my second year undergraduate lectures. About 70% of cell surface and extracellular matrix proteins are built at least partly from this simple domain structure. Like other superfolds such as the TIM barrel and globin these are protein structures with no sequence homology that are found in large numbers across biology and partly explain why there seems to be a limit to the number of different protein folds (Ref 2). But that is a story for another day.
The immunoglobulin-like fold is found in immunoglobulins (surprise, surprise), MHC, fibronectin, many surface receptors such as EGF receptors etc. etc. It’s also found in the largest protein known, Titin, which stops our muscles over extending when they are stretched. Now it turns up in this Caf1, which is beginning to nag me constantly with the thought that surely I could make things from this amazing polymer. So, off I go to the BBSRC with Mark Birch as co-investigator. With Liz Mitchell we had just shown that we could induce bone formation by growing osteoblasts on surfaces coated with engineered small proteins containing sequences from Bone morphogenic protein 2 and osteopontin (Ref 3). I said I could modify Caf1 to do the same and we could form it into hydrogels and grow 3D bone amongst other tissues – what could possibly go wrong. Funnily, the BBSRC did not agree and forbade any similar grant from darkening their doors for seven generations or something that sounded like that.
Try, try and try again
The project, such as it was, retreated to lick its wounds. At least my lack of citations meant that no one else in the world was likely to be working on it but neither it seemed was anyone interested in funding it. Salvation came in two forms. One was the MRC Industrial Collaboration studentship scheme, which enabled us to apply for money with Orla Protein Technologies Ltd, a spinout company I had co founded with Dale Athey in 2002. Orla sells engineered protein surface coatings such as those we used in the BMP/Osteopontin paper and very good they are too, try them yourselves at www.orlaproteins.com!
The second bit of luck was that Ana Roque applied for the studentship. She is an amazingly tenacious and hardworking scientist who single handedly made the Caf1 project work. First she tackled the ridiculously large Caf1 plasmid and won. We then had a manageable system that could produce mutant Caf1 in good amounts.

Immunoelectron microscopy of Y. pestis showing macrophage resistant hydrogel capsule From Du Y et al. Infect. Immun. 2002;70:1453-1460
She then showed that cells were not keen on growing on Caf1 surfaces. Not good. What turned it around was that by simply adding an RGDS motif we could make Caf1 act like fibronectin with cells sticking to it like last night’s pigs trotters. Caf1 makes a macrophage resistant hydrogel capsule around the Y. pestis cell that prevents interaction with cells (see figure). Ana had thus shown that it keeps its non-adhesive properties in vitro… whoo-hoo!. Thus, Caf1 is a non-stick, tough, flexible, polymer – all things very difficult to build into a protein by design. It can be genetically modified to imitate at least one Extra Cellular Matrix (ECM) protein and physically resembles many more. Ana then showed that we could cross-link it into hydrogels and her pièce de résistance was to show that we could make mixed polymers by expressing differently modified domains in the same cell and letting them mix randomly in the emerging polymer. This was published in Advanced Materials (Ref 4), which will do nicely as one of my REF papers although I am not waiting for the citations to roll in any time soon.

Ana returned to Portugal and the Caf1 project was halted again. We got some short term funding to pay Helen Waller’s salary to make more protein and more mutants.
 In November 2013 the Technology Strategy Board (TSB – now Innovate UK) announced the Industrial Biotechnology Catalyst with £20M in the first round. Although I have been involved with the bioprocessing industry for many years I did not think the Catalyst was for me, as it seemed to be either biopharmaceuticals or bioenergy. However, I read the outline for the feasibility awards and it seemed to fit Caf1. I was thinking of a post doc for three years etc but the award size started at £2M, so some imagination was required. This led to the current project. Protein engineering to design and produce new polymers will be done in my laboratory. This will be done by Helen Waller and a new post-doc. David Fulton will employ another post-doc to make our cross-linked hydrogels smart and responsive to a range of stimuli like temperature, light and pH. Matt German will use impressive kit like his Atomic Force Microscope (AFM) to measure the material properties of the gels whilst Nick Reynolds will continue our work on wound care. Last, but not least, ICaMB blog’s very own Neil Perkins will work with another new post doc to understand how cells respond to and remodel the different materials. Neil’s suggestion that his qualification was that he was the first person I met that day is unfounded, the truth is that his title of Prof Gene Expression and Signalling fitted the number of words left in that section of the form (I maintain that the previous 2 people Jeremy had bumped into that day had turned him down – Neil).
In November 2013 the Technology Strategy Board (TSB – now Innovate UK) announced the Industrial Biotechnology Catalyst with £20M in the first round. Although I have been involved with the bioprocessing industry for many years I did not think the Catalyst was for me, as it seemed to be either biopharmaceuticals or bioenergy. However, I read the outline for the feasibility awards and it seemed to fit Caf1. I was thinking of a post doc for three years etc but the award size started at £2M, so some imagination was required. This led to the current project. Protein engineering to design and produce new polymers will be done in my laboratory. This will be done by Helen Waller and a new post-doc. David Fulton will employ another post-doc to make our cross-linked hydrogels smart and responsive to a range of stimuli like temperature, light and pH. Matt German will use impressive kit like his Atomic Force Microscope (AFM) to measure the material properties of the gels whilst Nick Reynolds will continue our work on wound care. Last, but not least, ICaMB blog’s very own Neil Perkins will work with another new post doc to understand how cells respond to and remodel the different materials. Neil’s suggestion that his qualification was that he was the first person I met that day is unfounded, the truth is that his title of Prof Gene Expression and Signalling fitted the number of words left in that section of the form (I maintain that the previous 2 people Jeremy had bumped into that day had turned him down – Neil).
To boldly go…
So, we have five years to turn Caf1 into a range of 3D cell culture products. By exploiting bacterial production we hope to reduce costs and provide bulk materials at affordable prices for a series of applications. These will include research tools for cell culture, better culture conditions for industrial processes, tissue and regenerative engineering materials and many more. I have produced products in the past and it is very different to our normal hypothesis driven discovery research. In discovering things about nature you generally have observed something and then set out to find out why it’s like that or how it works. In product invention you never know if it will ever work because, by definition, no one knows (and that is particularly true of protein engineering!). The Industrial Biotechnology Catalyst money is aimed at expanding the UK biotechnology industry and, in taking the money, we must be aware that quite justifiably the tax payer wants to see a return on the investment. Ultimately we hope we can create jobs in a new area based upon this project. What’s more, collaborating across disciplines to invent amazing new materials and use them to cure disease is a great way to spend ones working life.
[1] Soliakov A, Harris JR, Watkinson A, Lakey JH. The structure of Yersinia pestis Caf1 polymer in free and adjuvant bound states. Vaccine. 2010;28:5746-54.
[2] Orengo CA, Jones DT, Thornton JM. Protein superfamilies and domain superfolds. Nature. 1994;372:631-4.
[3] Mitchell EA, Chaffey BT, McCaskie AW, Lakey JH, Birch MA. Controlled spatial and conformational display of immobilised bone morphogenetic protein-2 and osteopontin signalling motifs regulates osteoblast adhesion and differentiation in vitro. BMC Biology. 2010;8:57.
[4] Roque AI, Soliakov A, Birch MA, Philips SR, Shah DS, Lakey JH. Reversible Non-Stick Behaviour of a Bacterial Protein Polymer Provides a Tuneable Molecular Mimic for Cell and Tissue Engineering. Advanced Materials. 2014;26:2704-9.
 In a recent issue of Science, the discovery of a key mechanism for bacterial cell division was reported. This work was carried out by Dr Seamus Holden’s lab (ICaMB) in collaboration with Professor Cees Dekker (TU Delft), Professor Yves Brun (Indiana University), Professor Mike VanNieuwenhze (Indiana University) and Professor Ethan Garner (Harvard University). Here, Seamus tells us about this discovery and what its implications could be for antimicrobial research.
In a recent issue of Science, the discovery of a key mechanism for bacterial cell division was reported. This work was carried out by Dr Seamus Holden’s lab (ICaMB) in collaboration with Professor Cees Dekker (TU Delft), Professor Yves Brun (Indiana University), Professor Mike VanNieuwenhze (Indiana University) and Professor Ethan Garner (Harvard University). Here, Seamus tells us about this discovery and what its implications could be for antimicrobial research.