Blogs are now a widespread science communication tool, with many researchers taking to the blogosphere to discuss the latest scientific discoveries, explain the basic concepts in their research field to a wide audience or just talk about science and scientists. Our 3rd year bioscience students have done just that, and this week we have a guest post prepared by them.
by Antisense Science
Antisense Science is a science blog founded by a group of 3rd year bio-scientists from Newcastle University. As a team, we recognise that science is not as accessible to the general public as it should be. We therefore make it our primary aim to translate complex scientific principles and research articles that interest us personally, into topical, thought provoking blogs accessible to everyone.
Our project is small but our ambition is big! Since our founding in October 2013 we have published 58 articles covering psychoactive baths salts, human evolution and the neurobiology of love, to name a few, and with a growing following (we’ve had over 6000 hits since inception) we were thrilled to be given the opportunity to guest post on ICaMBlog. As Newcastle University students, our interest in research was stirred by the professors at Newcastle University, including those who founded this blog. With planned guest posts focusing on research by Prof Rick Lewis among others, maybe YOU will feature on Antisense Science in the near future! We foresee (we hope!) that our blog can form a bridge between researchers here at Newcastle and the student body, raising awareness of what is actually being discovered right under our noses. By forming mutually beneficial collaborations, we hope to diversify and grow our following and expose our current readers to a continual stream of stimulating articles which never fail to pique the interest of the curious.

Meet the Antisense Science team
Currently, we have a total of 7 writers, all of whom enjoy sharing the intrigue of the latest developments in science, from biochemistry to microbiology (and even physics) and we have no plans of stopping. Although many of us will be moving on from our BScs to ever greater things, Antisense Science will remain and we are even in the process of recruiting further up and coming bio-scientists as writers (keep your eyes peeled for blogs from first year students Bethany Lumborg and Lucy Gee, as well as our fellow third year Emily Lawson and a multitude of guest posters from across the student body). The future looks bright and we’re very glad with the progress we have made thus far!
For an example of what we do, here is an article on depression written by our very own Joe Sheppard. We hope you enjoy it! If you ever want to be involved in any of our projects feel free to drop us a message. We also have Facebook (www.facebook.com/antisensescience) and Twitter (@AntisenseSci) so there are plenty of ways to keep in the loop.
The Confounding Contradictions of Depression
If you currently have or have had depression then you may already be able to tell your MAOIs from your SSRIs, but if you haven’t then what you read here might actually help. Knowledge is power and I believe learning a little something about depression could contribute a bit of control to an otherwise daunting and often underestimated medical condition.
Depression is perhaps the ultimate “common complex disorder”. Unlike pathogens or mutations that affect physical body tissue, depression is a condition that alters the very consciousness and emotional state of an individual making it a truly unique affliction. Throughout the course of our lives 1 in 5 of us will experience depression or anxiety of some kind, yet the majority of people conceive depression simply as a disease of “sadness” when the truth is much more complex. “Anhedonia“, an inability to feel joy in anything and “congruent memory bias”, the inability to remember or altered recall of specific memories, are extremely common cognitive behaviours in depression that we often inflict upon ourselves on a day to day basis.
It may surprise you to know that modern science can say with little certainty what neuro-physiological changes initiate depression, and linking those that we do think are involved to the broad psychiatric manifestations seen in cases of depression is even harder as human experience and consciousness is beyond the understanding of molecular neuroscience (and by extension, definitely me). But from the murky depths of our own minds patterns do emerge, and as such there are a few good theories out there.
Rather confusingly the best fitting theory of depression is actually based on the drugs WE ALREADY USE to treat it, not a common theme in medicine, I might add. “The monoamine theory of depression” states that depressed brains have reduced signalling between neurons via a group of specific chemical neurotransmitters called monoamines. Two in particular called 5-hydroxytryptophan (hereafter referred to as serotonin) and dopamine are released into a synapse to induce electrical signals between neurons in the midbrain. These two neurotransmitters and the resulting electrical signals are most notably perceived as feelings of joy, euphoria, reward and attention. And there is some evidence to back this up: depletion of tryptophan, an amino acid essential for serotonin synthesis in the brain, caused mood congruent memory bias, and altered reward-related behaviours. Biochemical evidence exists too, abnormalities of the protein that binds serotonin in the brain called the 1A receptor have been noted in multiple brain areas of major depressive disorder (MDD) patients. Why does serotonin decline in patients with depression? Well, search my pockets, you will find no answers.
Rather reassuringly this hasn’t stopped treatment of depression at all, and several drugs for which the theory is named are all targeted at increasing serotonin, and so good feelings, within the brain. The so named selective serotonin re-uptake inhibitors (those “SSRIs” I snuck in earlier, such as fluoxetine and sertraline) are the most prescribed group of antidepressants and work in a way best aided pictographically:
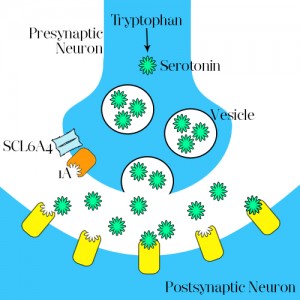
Schematic representation of neuron activity
Serotonin is synthesised in the presynaptic neuron from tryptophan, from here it is packaged into vesicles and upon nerve impulse firing (see previous article “ shedding light on neural networks”) is released in the synaptic cleft (the space between neurons). Serotonin then binds to receptors on the postsynaptic neuron, stimulating a similar nerve impulse. However, serotonin is also able to control its own release: By binding to the 1A receptor on the presynaptic neuron it prevents continued release of serotonin, allowing the proposed channel protein SCL6A4 to re-absorb serotonin in the presynaptic neuron to be destroyed.
This is where SSRIs come in. Believed to bind to SCL6A4 and prevent the re-absorption of serotonin, it allows serotonin to remain in the synaptic cleft for longer and therefore stimulate nerve impulses in the post synaptic neuron for longer, and so increase the degree of monoamine signalling.
Let’s not forget our friend dopamine:
Dopamine too is a monoamine consistently found reduced in the blood of patients with depression, as a result of decreased synthesis and degradation in the brains of these patients. Neurons that signal using dopamine (as opposed to serotonin) are found in a region of the brain called the substantia nigra that degenerates during Parkinson’s disease. Interestingly the motor impairment (shaking) in this disease is often preceeded by major depressive disorder in 50% of Parkinson’s cases!
This brings me quite nicely to my final point. Since so little is known about the basis of psychiatric disorders their treatment has been almost purely symptomatic for the last 60 years. Thomas Insel, director of the US National Institute for Mental Health was quoted in 2013 as saying “In the rest of medicine, this would be equivalent to creating diagnostic systems based on the nature of chest pain, or the quality of fever.” This was following the release of the 5th edition of the diagnostic and statistical manual of mental disorders (DSM-5), that diagnoses psychiatric conditions based on common symptoms presented by each condition. Despite the strong agreement with Insel by many psychiatrists that this method is outdated, the shift to a more objective and molecular diagnostic is years away given the outstanding complexity of these diseases. But keep the hope, recent booms in neuroscience research are a sure step towards a less archaic means of treating depression and other mental disorders.
A new approach spearheaded by the same Thomas Insel called “Research Domain Criteria” or RDoC is already doing just that by utilizing genomic sequencing technology, fMRI imaging techniques and cognitive science to develop an entirely new platform for diagnosis based on new data being attained all the time, rather than the laboured DSM-5 classification. While still in its infancy this new approach aims to start looking at broader groups for diagnosis rather than classification of different disorders by the symptoms they present. For instance, by looking at a group of patients with different disorders but all experiencing anhedonia will not only allow a greater insight into a unifying cause for these symptoms, but also speed up treatments in the future.
Of course, there are always two sides to each argument and depression does in some sense stand alone from other psychiatric disorders such as schizophrenia in that it imparts a greater emotional influence on the sufferer – if “precision medicine” were able to prescribe a pill for the treatment of emotional conditions, would you want it to? Of course this is a quandary we won’t face for some time, but worth a thought.
J.
Some interesting but by no means comprehensive reviews on depression, its far too huge for 3 articles!
Hasler G (2010). Pathophysiology of depression: do we have any solid evidence of interest to clinicians? World psychiatry : official journal of the World Psychiatric Association (WPA), 9 (3), 155-61 PMID: 20975857
Martinowich K, Manji H, & Lu B (2007). New insights into BDNF function in depression and anxiety. Nature neuroscience, 10 (9), 1089-93 PMID: 17726474
Frances, A. (2013) One manual shouldn’t dictate US mental health research
(Accessed: 07/01/14).